Publications
For a full list see below or go to
Google Scholar,
Research Gate,
Orcid
Highlights
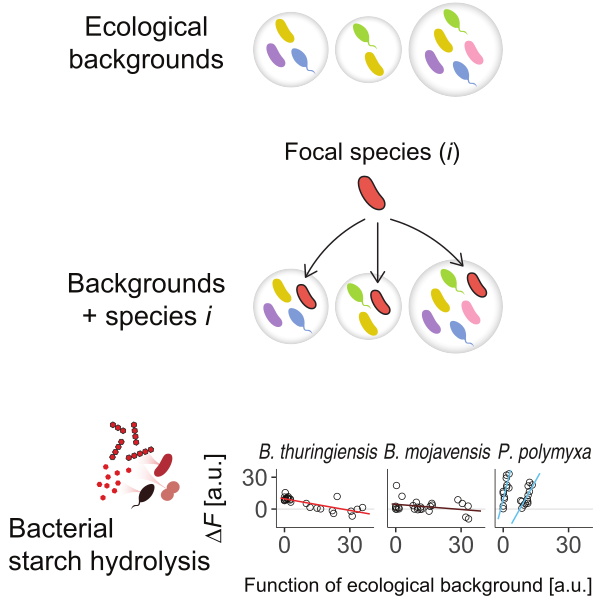
We found that global epistasis – an emergent phenomenon in fitness landscapes – can be applied to predict the function of microbial communities.
Diaz-Colunga, J*; A. Skwara*, K., J.C.C. Vila, D. Bajić#, A. Sanchez#
Cell. 187(12):3108-3119.e30. 2024
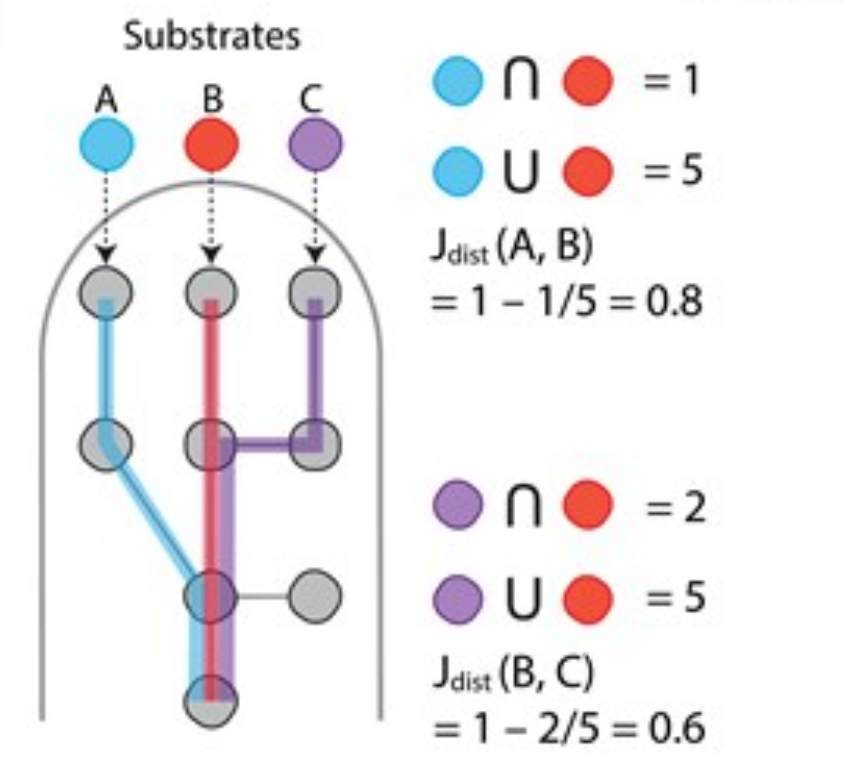
We explored the evolvability of microbial resource use hierarchies using metabolic modeling.
Takano, S#;, J.C.C. Vila, R. Miyazaki, Á. Sánchez, D. Bajić#
Mol. Biol. Evol.. 40(9):msad187. 2023
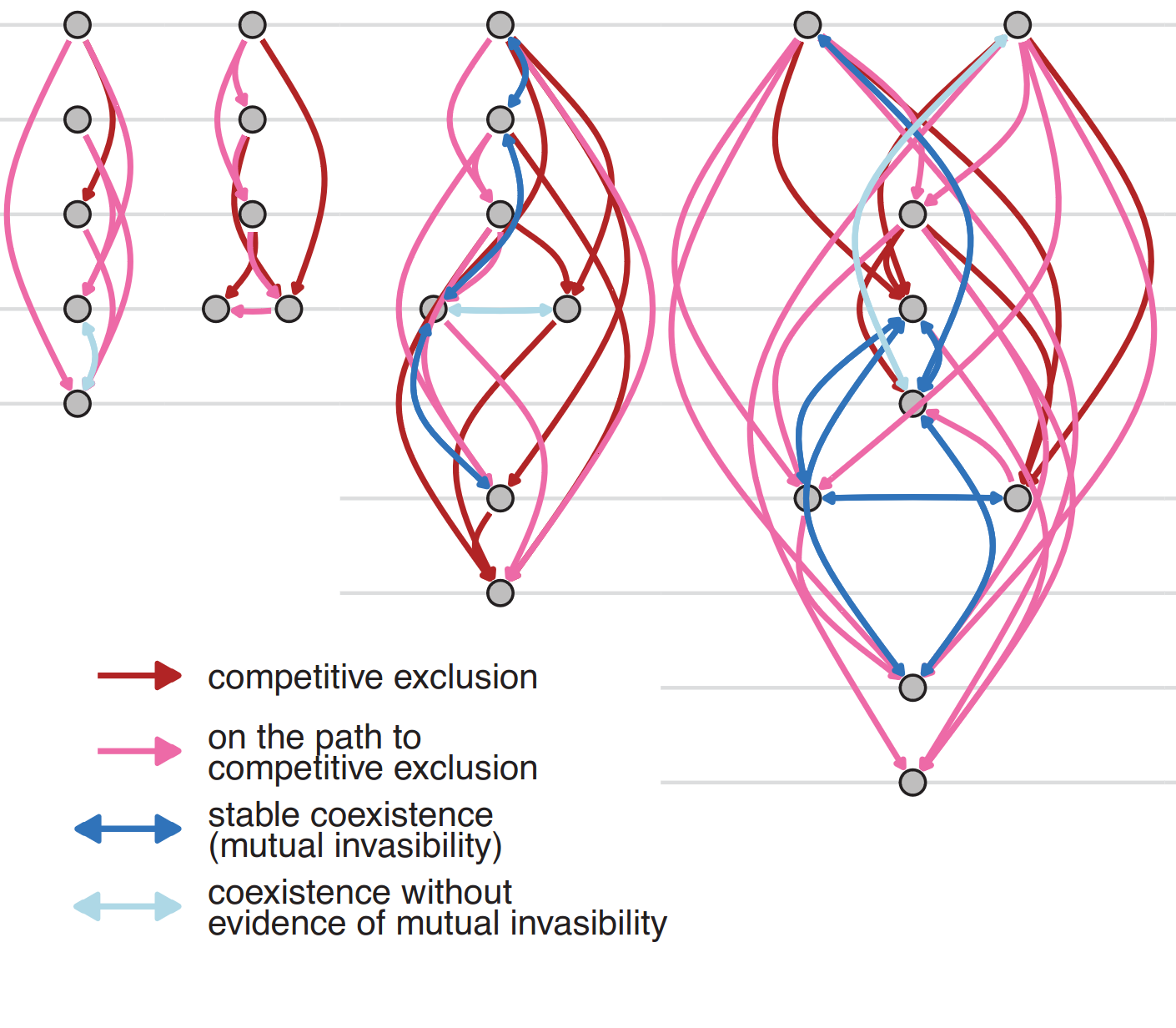
In diverse microbial ecosystems, pairs of species often cannot coexist in isolation from the larger community context.
Chang, C-Y.#; D. Bajić#, J.C.C. Vila, S. Estrela, A. Sanchez#
Science. 381 (6655), 343-348. 2023
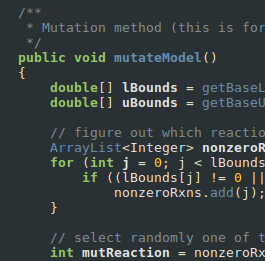
A state-of-the-art platform for genome-scale metabolic modeling of multi-species communities.
Dukovski*, I; D. Bajić*,J.M. Chacon*, M. Quintin*, J.C.C. Vila, S. Sulheim, A.R. Pacheco, D.B.Bernstein, W.J. Riehl, K.S. Korolev, A. Sánchez, W.R. Harcombe, D. Segrè
Nature Protocols. 16:5030–5082. 2021
Full List
[23] Diaz-Colunga, J*; A. Skwara*, K., J.C.C. Vila, D. Bajić#, A. Sanchez#
Global epistasis and the emergence of function in microbial consortia
2024. Cell. 187(12):3108-3119.e30
-
[22] Vila, J.C.C.;, J. Goldford, S. Estrela, D. Bajic, A. Sanchez-Gorostiaga, A. Damian-Serrano, N. Lu, R. Marsland III, M. Rebolleda-Gomez, P. Mehta, A. Sanchez
Metabolic similarity and the predictability of microbial community assembly
2024. bioRxiv. 10.1101/2023.10.25.564019
-
[21] Takano, S#;, J.C.C. Vila, R. Miyazaki, Á. Sánchez, D. Bajić#
The architecture of metabolic networks constrains the evolution of microbial resource hierarchies
2023. Molecular Biology and Evolution. 40(9):msad187
-
[20] Scott Jr, W.T; S. Benito-Vaquerizo, J. Zimmermann, D. Bajić, A. Heinken, M. Suarez-Diez, P.J. Schaap
A structured evaluation of genome-scale constraint-based modeling tools for microbial consortia
2023. PLoS Computational Biology. 19(8):e1011363
-
[19] Chang, C-Y.#; D. Bajić#, J.C.C. Vila, S. Estrela, A. Sanchez#
Emergent coexistence in multispecies microbial communities
2023. Science. 381 (6655), 343-348
-
[18] Diaz-Colunga, J*; A. Skwara*, K. Gowda, R. Diaz-UriarteJ.C.C. Vila, M. Tikhonov, D. Bajić#, A. Sanchez#
Global epistasis on fitness landscapes
2023. Philosophical Transactions of the Royal Society B. 378(1877):20220053
-
[17] Sanchez, A.; D. Bajić, J. Diaz-Colunga, A. Skwara, J.C.C. Vila, S. Kuehn
The community-function landscape of microbial consortia
2023. Cell Systems. 14(1):122-134
-
[16] Estrela, S.*#; J.C.C. Vila, N. Lu, D. Bajić, M. Rebolleda-Gomez, C-Y. Chang, A. Sánchez #,
Functional attractors in microbial community assembly
2022. Cell Systems. 13(1):29-42
bioRxiv - -
[15] Dukovski*, I; D. Bajić*,J.M. Chacon*, M. Quintin*, J.C.C. Vila, S. Sulheim, A.R. Pacheco, D.B.Bernstein, W.J. Riehl, K.S. Korolev, A. Sánchez, W.R. Harcombe, D. Segrè
A metabolic modeling platform for the computation of microbial ecosystems in time and space (COMETS)
2021. Nature Protocols. 16:5030–5082
arXiv - -
[14] Alonso-Lavin A.J; D. Bajić, J.F. Poyatos
Tolerance to NADH/NAD+ imbalance anticipates aging and anti-aging interventions
2021. iScience. 102697
- -
[13] Bajić D #; M. Rebolleda-Gomez, M.M. Muñoz, A. Sánchez #
The macroevolutionary consequences of niche construction in microbial metabolism
2021. Frontiers in Microbiology.
-
[12] de Oliveira Lino, F.S.; D. Bajić, J.C.C. Vila, A. Sánchez, M.O.A. Sommer
Complex yeast–bacteria interactions affect the yield of industrial ethanol fermentation
2021. Nature communications. 12(1):1-12
- -
[11] Peralta-Marzal L.N.#; N. Prince, D. Bajić, L. Roussin, L. Naudon, S. Rabot, J. Garssen, A.D. Kraneveld, P. Pérez-Pardo #
The Impact of Gut Microbiota-Derived Metabolites in Autism Spectrum Disorders
2021. International Journal of Molecular Sciences. 22(18):10052
- -
[10] Kovács*, K.; Z. Farkas*, D. Bajić*, D. Kalapis, A. Daraba, K. Almási, B. Kintses, Z. Bódi, R. Notebaart, J.F. Poyatos, P. Kemmeren, F.C.P. Holstege, C. Pál, B. Papp,
Suboptimal global transcriptional response increases the harmful effects of loss-of-function mutations
2020. Molecular Biology and Evolution. 38(3):1137–1150
- -
[9] Bajić, D. and A. Sánchez
The ecology and evolution of microbial metabolic strategies
2020. Current Opinion in Biotechnology. 62:123-128
- -
[8] Chang, C-Y* and M.L. Osborne*, D. Bajić, A. Sánchez
Artificially selecting bacterial communities using propagule strategies,
2020. Evolution. 74(10):2392-2403
- -
[7] Sánchez-Gorostiaga*, A; D. Bajić*, M.L. Osborne, J.F. Poyatos, and A. Sánchez
High-order interactions distort the functional landscape of microbial consortia,
2019. PLoS Biology. 17(12): e3000550
- -
[6] D. Bajić*#, J.C.C. Vila*, Z. Blount, A. Sánchez #
On the deformability of an empirical fitness landscape by microbial evolution
2018. Proceedings of the National Academy of Sciences. 115 (44): 11286–11291
- -
[5] Goldford*, J.E.; N. Lu*, D. Bajić, S. Estrela, M. Tikhonov, A. Sánchez-Gorostiaga, D. Segrè, P. Mehta #, A. Sánchez #,
Emergent simplicity in microbial community assembly,
2018. Science. 361(6401):469-474
- -
[4] G. Rodrigo, D. Bajić, I. Elola, J.F. Poyatos
Deconstructing a multiple antibiotic resistance regulation through the quantification of its input function
2017. NPJ systems biology and applications. 3(1):1–9
- -
[3] G. Rodrigo, D. Bajić, I. Elola, J.F. Poyatos
Antagonistic autoregulation speeds up a homogeneous response in Escherichia coli
2016. Scientific Reports. 6 (1): 1–10
- -
[2] Bajić, D; C. Moreno-Fenoll, J.F. Poyatos
Rewiring of genetic networks in response to modification of genetic background},
2014. Genome Biology and Evolution. 6(12):3267–3280
- -
[1] D. Bajić, J. Poyatos
Balancing noise and plasticity in eukaryotic gene expression
2012. Proceedings of the National Academy of Sciences. 13 (1): 1–12
- -