Research
We seek to predictively understand how microbial communities assemble, function, and respond to environmental change. Below is a glimpse of some of the questions we are currently interested in answering.

Understanding the metabolic principles of microbial community assembly. The chemical composition of the environment is a critical driver of the species composition of microbial communities. We work towards understanding quantitatively how the composition of microbial communities is determined by their metabolic environment, and how it responds to metabolic perturbation. To this end, we combine experiments in which we assemble of model microbial communities (both in high-throughput and in bioreactors, where we collaborate with the UNLOCK facility), and modeling. Achieving this goal could pave the way to precisely engineering mirobial community composition through the manipulation of their metabolic environment.
[Science 2023] [Cell Systems 2022]
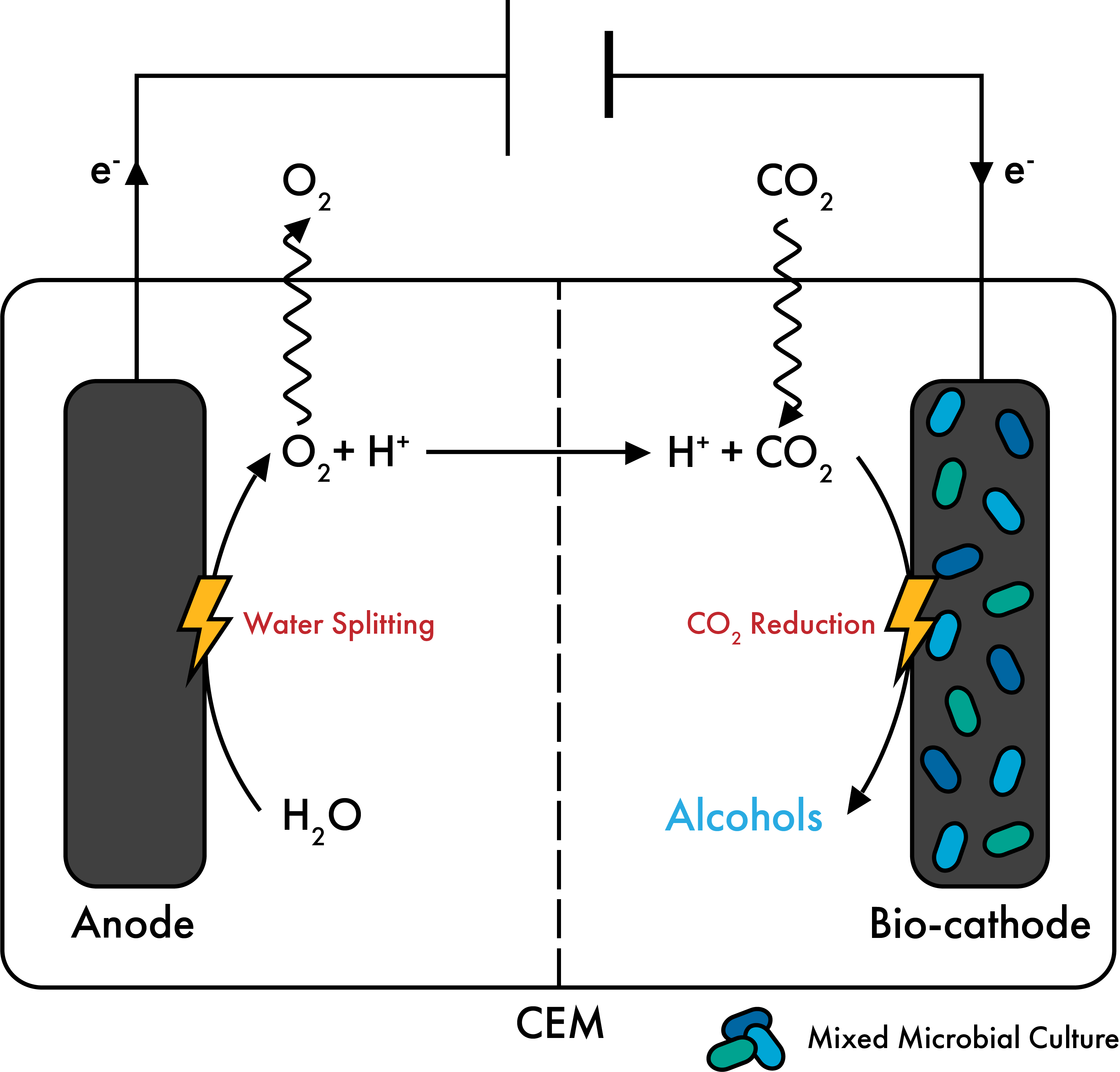 e-COMM: Understanding and engineering electrosynthetic communities.
As part of the Zero Emission Biotechnology Program at TU Delft, we are steering our expertise to study biofilm microbial communities transforming green electricity and CO2 to valuable products. This project is a collaboration with the groups of Ludovic Jourdin and Jean-Marc Daran in our department.
e-COMM: Understanding and engineering electrosynthetic communities.
As part of the Zero Emission Biotechnology Program at TU Delft, we are steering our expertise to study biofilm microbial communities transforming green electricity and CO2 to valuable products. This project is a collaboration with the groups of Ludovic Jourdin and Jean-Marc Daran in our department.
© Image courtesy of Marika Zegers.
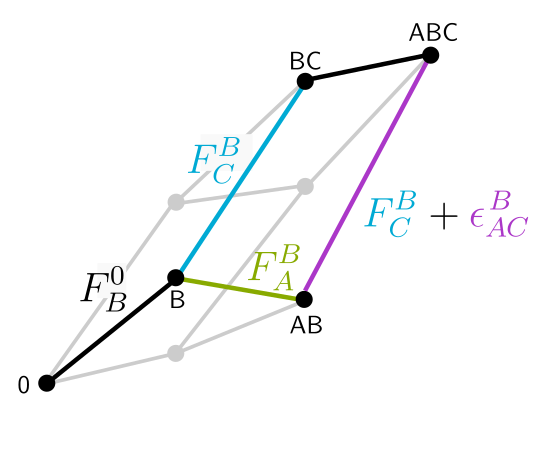
Engineering microbiome function through its emergent properties Understanding in detal how microbial species interact within communities to collectively deliver a function represents a colossal task. We collaborate with Alvaro Sanchez, Juan Diaz-Colunga, and others, to take an alternative, statistical approach. Instead of aiming to understand first all the details, we take a shortcut and “start from the roof” by looking for emergent patterns that can help us engineer communities with a desired output, without necessarily understanding the underlying mechanisms. We also apply these methods in collaboration with researchers in different fields to help them engineer microbial communities with interesting functions, for example or those helping crops grow and resist pathogens (Ivica Dimkić) or those fermentating soymilk (Nemanja Stanisavljević and Nemanja Kljajević).
[Cell, 2024] [Cell Syst., 2023] [Proc.R. Soc. B, 2023] [Nat. Comm,, 2021]
New methods: building digital twins of microbial communities. Guided by the principle “if you can’t build it, you don’t understand it”, we are building mechanistic, metagenome-scale microbial community metabolic models. We are part of the developer team of COMETS, a software platform for the “Computation of microbial ecosystems in time and space”, in collaboration with Daniel Segrè and Ilija Dukovski.
[PLoS Comp. Biol. 2023]
[Nature Protocols 2021]
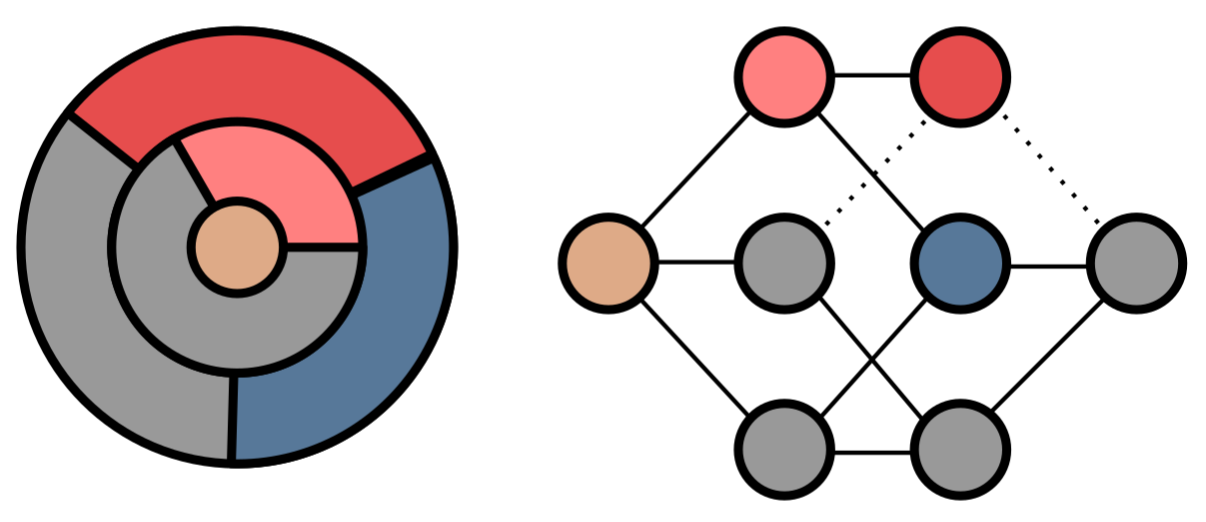 Using metabolic models to understand ecology and evolution.
We are interested in a broad range of topics in microbial ecology and evolution. We address these use metabolic models as a workhorse to explore empirically calibrated, realistic genotype-phenotype-ecology maps. We are interested in a broad range of topics in microbial ecology and evolution. For example, we are currently collaborating with Maria Rebolleda-Gómez to study how microbial metabolisms innovate during evolution.
Using metabolic models to understand ecology and evolution.
We are interested in a broad range of topics in microbial ecology and evolution. We address these use metabolic models as a workhorse to explore empirically calibrated, realistic genotype-phenotype-ecology maps. We are interested in a broad range of topics in microbial ecology and evolution. For example, we are currently collaborating with Maria Rebolleda-Gómez to study how microbial metabolisms innovate during evolution.
[Mol. Biol. Evol. 2023] [Current Opinion in Biotechnology 2021] [Frontiers in Microbiology 2021]